I am Distinguished Research Fellow at the Biodiversity Research Center, Academia Sinica (BRCAS) in Taipei. Before moving back to Taiwan in 2013, I was the James Watson Professor, Department of Ecology and Evolution, University of Chicago.
I received my PhD in applied mathematics, but have been working on evolutionary biology since my graduate study. My research interests are population genetics, bioinformatics, molecular evolution, and genomic evolution.
期刊論文
- Chen HJ, A Sawasdee, YL Lin, MY Chiang, HY Chang, WH Li*, CS Wang*, 2024, “Reverse Mutations in Pigmentation Induced by Sodium Azide in the IR64 Rice Variety”, Current Issues in Molecular Biology, 46(12), 13328-13346. (SCIE) (IF: 3.1; SCI ranking: 60.7%)
- Pi HW, Chiang YR, Li WH*, 2024, “Mapping Geological Events and Nitrogen Fixation Evolution Onto the Timetree of the Evolution of Nitrogen-Fixation Genes.”, Molecular biology and evolution, 41(2), msae023. (SCIE) (IF: 10.7; SCI ranking: 8.8%,7.5%,5.2%)
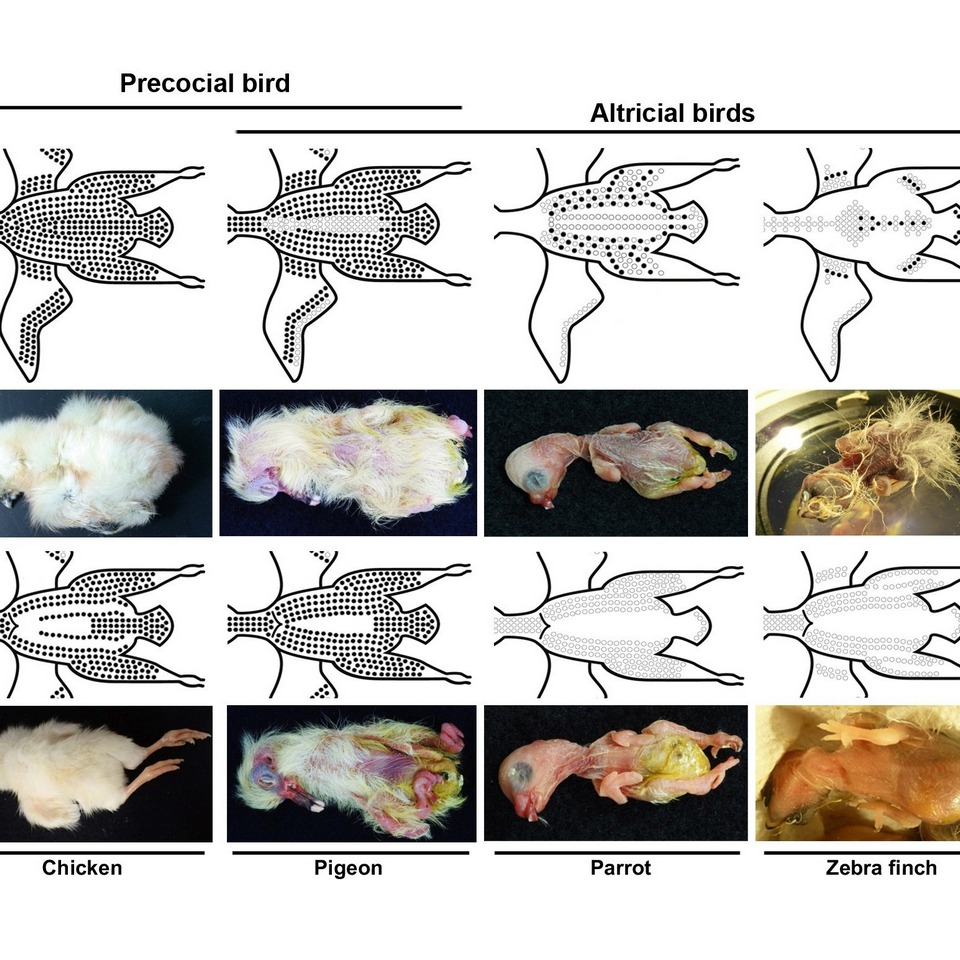
- Chen CK, Chang YM, Jiang TX, Yue Z, Liu TY, Lu J, Yu Z, Lin JJ, Vu TD, Huang TY, Harn HI, Ng CS, Wu P, Chuong CM, Li WH*, 2024, “Conserved regulatory switches for the transition from natal down to juvenile feather in birds.”, Nature communications, 15(1), 4174. (SCIE) (IF: 16.6; SCI ranking: 8.2%)
- Wu CC, Hsieh KT, Yeh SY, Lu YT, Chen LJ, Ku MSB, Li WH*, 2023, “Simultaneous detection of miRNA and mRNA at the single-cell level in plant tissues.”, Plant biotechnology journal, 21, 36-149. (SCIE) (IF: 13.263; SCI ranking: 5%,2.1%)
- Tsai DY, Chen JJ, Su PC, Liu IM, Yeh SH, Chen CK, Cheng HC, Chen CF, Li WH, Ng CS, 2023, “Chicken HOXC8 and HOXC10 genes may play a role in the altered skull morphology associated with the Crest phenotype.”, Journal of experimental zoology. Part B, Molecular and developmental evolution, 340(6), 392-402. (SCIE) (IF: 5.511; SCI ranking: 35.7%,20.8%)
- Tseng KC, Huang HT, Huang SN, Yang FY, Li WH, Nan FH, Lin YJ, 2023, “Lactobacillus plantarum isolated from kefir enhances immune responses and survival of white shrimp (Penaeus vannamei) challenged with Vibrio alginolyticus.”, Fish & shellfish immunology, 135, 108661. (SCIE) (IF: 4.622; SCI ranking: 11.1%,51.5%,9.7%,4.1%)
- Bhattacharjee MJ, Bhattacharya A, Kashyap B, Taw MJ, Li WH*, Mukherjee AK, Khan MR, 2023, “Genome analysis of SARS-CoV-2 isolates from a population reveals the rapid selective sweep of a haplotype carrying many pre-existing and new mutations.”, Virology journal, 20(1), 201. (SCIE) (IF: 5.916; SCI ranking: 35.1%)
- Hsieh KT, Wu CC, Lee SJ, Chen YH, Shiue SY, Liao YC, Liu SH, Wang IW, Tseng CS, Li WH, Wang CS, Chen LJ, 2023, “Rice GA3ox1 modulates pollen starch granule accumulation and pollen wall development.”, PloS one, 18(10), e0292400. (SCIE) (IF: 3.752; SCI ranking: 39.2%)
- Kobayashi N, Dang TA, Pham KTM, Gomez Luciano LB, Van Vu B, Izumitsu K, Shimizu M, Ikeda KI, Li WH*, Nakayashiki H, 2023, “Horizontally Transferred DNA in the Genome of the Fungus Pyricularia oryzae is Associated With Repressive Histone Modifications.”, Molecular biology and evolution, 40(9), msad186. (SCIE) (IF: 8.8; SCI ranking: 13.1%,9.6%,8.5%)
- Pi HW, Lin JJ, Chen CA, Wang PH, Chiang YR, Huang CC, Young CC, Li WH*, 2022, “Origin and Evolution of Nitrogen Fixation in Prokaryotes”, Molecular biology and evolution, 39(9), msac181. (SCIE) (IF: 8.8; SCI ranking: 13.1%,9.6%,8.5%)
- Samuel SY, Wang HD, Huang MY, Cheng YS, Chen JR, Li WH, Chang JJ, 2022, “Safety Assessment of 3S, 3'S Astaxanthin Derived from Metabolically Engineered K. marxianus.”, Antioxidants, 11(11), 1-16. (SCIE) (IF: 7.675; SCI ranking: 16.8%,6.3%,8.3%)
- Lo KL, Chen YN, Chiang MY, Chen MC, Panibe JP, Chiu CC, Liu LW, Chen LJ, Chen CW, Li WH, Wang CS, 2022, “Two genomic regions of a sodium azide induced rice mutant confer broad-spectrum and durable resistance to blast disease.”, Rice, 15(1), 2. (SCIE) (IF: 5.638; SCI ranking: 12.2%)
- Liu WY, Yu CP, Chang CK, Chen HJ, Li MY, Chen YH, Shiu SH, Ku MSB, Tu SL, Lu MJ, Li WH*, 2022, “Regulators of early maize leaf development inferred from transcriptomes of laser capture microdissection (LCM)-isolated embryonic leaf cells.”, Proceedings of the National Academy of Sciences of the United States of America, 119(35), e2208795119. (SCIE) (IF: 12.779; SCI ranking: 12.2%)
- Panibe JP, Wang L, Lee YC, Wang CS, Li WH, 2022, “Identifying mutations in sd1, Pi54 and Pi-ta, and positively selected genes of TN1, the first semidwarf rice in Green Revolution.”, Botanical studies, 63(1), 9. (SCIE) (IF: 2.673; SCI ranking: 40.8%)
- Ng CS, Lai CK, Ke HM, Lee HH, Chen CF, Tang PC, Cheng HC, Lu MJ, Li WH, Tsai IJ, 2022, “Genome Assembly and Evolutionary Analysis of the Mandarin Duck Aix galericulata Reveal Strong Genome Conservation among Ducks”, Genome biology and evolution, 14(6), evac083. (SCIE) (IF: 4.065; SCI ranking: 32.7%,41.2%)
- Tusher TR, Chang JJ, Saunivalu MI, Wakasa S, Li WH, Huang CC, Inoue C, Chien MF, 2022, “Second-generation bioethanol production from phytomass after phytoremediation using recombinant bacteria-yeast co-culture”, Fuel, 326, 124975-124975. (SCIE) (IF: 8.035; SCI ranking: 24.4%,13.3%)
- Su-Ying Y, Lin HH, Chang YM, Chang YL, Chang CK, Huang YC, Ho YW, Lin CY, Zheng JZ, Jane WN, Ng CY, Lu MY, Lai IL, To KY, Li WH, Ku MSB, 2022, “Maize Golden2-like transcription factors boost rice chloroplast development, photosynthesis and grain yield.”, Plant physiology, 188(1), 442-459. (SCIE) (IF: 8.005; SCI ranking: 5%)
- Huang CF, Liu WY, Lu MYJ, Chen YH, Ku MSB, Li WH*, 2021, “Whole genome duplication facilitated the evolution of C4 photosynthesis in Gynandropsis gynandra”, Molecular biology and evolution, 38 (11) , 4715-4731. (SCIE) (IF: 8.8; SCI ranking: 13.1%,9.6%,8.5%)
- Ko SS, Li MJ, Ho YC, Yu CP, Yang TT, Lin YJ, Hsing HC, Chen TK, Jhong CM, Li WH, Sun-Ben Ku M, 2021, “Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development”, Journal of experimental botany, 72(13), 4888-4903. (SCIE) (IF: 7.378; SCI ranking: 6.3%)
- Yu CP, Kuo CH, Nelson CW, Chen CA, Soh ZT, Lin JJ, Hsiao RX, Chang CY, Li WH*, 2021, “Discovering unknown human and mouse transcription factor binding sites and their characteristics from ChIP-seq data.”, Proceedings of the National Academy of Sciences of the United States of America, 118(20), e2026754118. (SCIE) (IF: 12.779; SCI ranking: 12.2%)
- Joyraj Bhattacharjee M, Lin JJ, Chang CY, Chiou YT, Li TN, Tai CW, Shiu TF, Chen CA, Chou CY, Chakraborty P, Yuan Tseng Y, Hui-Ching Wang L, Li WH*, 2021, “Identifying primate ACE2 variants that confer resistance to SARS-CoV-2.”, Molecular biology and evolution, 38(7), 2715–2731. (SCIE) (IF: 8.8; SCI ranking: 13.1%,9.6%,8.5%)
- Liu HL, Wang CH, Chiang EI, Huang CC, Li WH*, 2021, “Tryptophan plays an important role in yeast's tolerance to isobutanol.”, Biotechnology for biofuels, 14(1), 200. (SCIE) (IF: 7.67; SCI ranking: 11.8%,26.1%)
- Panibe JP, Wang L, Li JY, Li MY, Lee YC, Wang CS, Ku MSB, Lu MY, Li WH*, 2021, “Chromosomal-level genome assembly of the semi-dwarf rice Taichung Native 1, an initiator of Green Revolution”, Genomics, 113(4), 2656-2674. (SCIE) (IF: 4.31; SCI ranking: 36.6%,33.9%)
- Liu Wen-Yu, Lin Hsin-Hung, Yu Chun-Ping, Chang Chao-Kang, Chen Hsiang-June, Lin Jinn-Jy, Lu Mei-Yeh Jade, Tu Shih-Long, Shiu Shin-Han, Wu Shu-Hsing, Ku Maurice S. B., Li Wen-Hsiung*, 2020, “Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth”, Proceedings of the National Academy of Sciences of the United States of America, 117(35), 21747-21756. (SCIE) (IF: 12.779; SCI ranking: 12.2%)
- Vu Trieu-Duc, Iwasaki Yuki, Shigenobu Shuji, Maruko Akiko, Oshima Kenshiro, Iioka Erica, Huang Chao-Li, Abe Takashi, Tamaki Satoshi, Lin Yi-Wen, Chen Chih-Kuan, Lu Mei-Yeh, Hojo Masaru, Wang Hao-Ven, Tzeng Shun-Fen, Huang Hao-Jen, Kanai Akio, Gojobori Takashi, Chiang Tzen-Yuh, Sun H. Sunny, Li Wen-Hsiung, Okada Norihiro, 2020, “Behavioral and brain- transcriptomic synchronization between the two opponents of a fighting pair of the fish Betta splendens”, PLoS Genetics, 16(6), e1008831. (SCIE) (IF: 6.02; SCI ranking: 15.3%)
- Lee MH, Hsu TL, Lin JJ, Lin YJ, Kao YY, Chang JJ, Li WH*, 2020, “Constructing a human complex type N-linked glycosylation pathway in Kluyveromyces marxianus.”, PloS one, 15(5), e0233492. (SCIE) (IF: 3.752; SCI ranking: 39.2%)
- Anandharaj M, Lin YJ, Rani RP, Nadendla EK, Ho MC, Huang CC, Cheng JF, Chang JJ, Li WH*, 2020, “Constructing a yeast to express the largest cellulosome complex on the cell surface.”, Proceedings of the National Academy of Sciences of the United States of America, 117(5), 2385-2394. (SCIE) (IF: 12.779; SCI ranking: 12.2%)
- Wang WF, Lu MJ, Cheng TR, Tang YC, Teng YC, Hwa TY, Chen YH, Li MY, Wu MH, Chuang PC, Jou R, Wong CH, Li WH*, 2020, “Genomic Analysis of Mycobacterium tuberculosis Isolates and Construction of a Beijing Lineage Reference Genome.”, Genome biology and evolution, 12(2), 3890-3905. (SCIE) (IF: 4.065; SCI ranking: 32.7%,41.2%)
Tel: +886-2-2787-2256
Lab Tel: +886-2-2787-2257
Email: whli gate.sinica.edu.tw
gate.sinica.edu.tw
 gate.sinica.edu.tw
gate.sinica.edu.twAddress: Interdisciplinary Research Building for Science and Technology A203